Program
Invited speakers
Topics and Program Overview
The program will be structured into three oral and one poster sessions. Each of the oral sessions will be started with an invited keynote lecture dealing with methodical aspects of metabolomics, transcriptomics and chemometric data evaluation. In these symposium topics, participants are invited to communicate their research and findings through oral or poster presentations. All abstracts submitted for consideration will undergo a review process by the scientific committee.
A poster prize will be awarded to the top poster presentation at the symposium. The winning poster will be voted by all participants. This is a great opportunity, especially for young scientists to showcase their work and receive recognition for their efforts.
Program
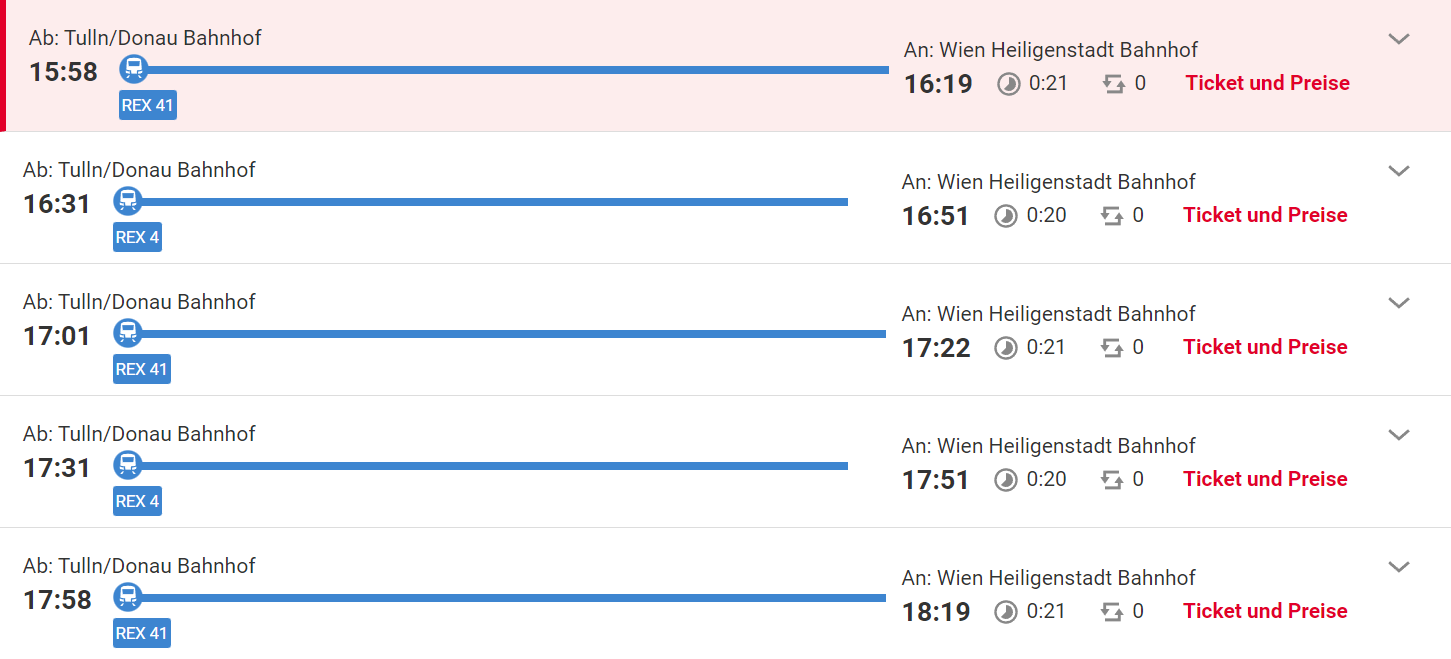
The final program of the Symposium is as follows:
Registration and welcome coffee
9:00 - 10:00
1st session
10:10 - 11:50
Chairs: Rainer Schuhmacher, Jonathan Samson
Keynote 10:10 - 10:50
Gerhard G. Thallinger
Reliable metabolite identification: From spectrum scoring to decision rules to deep learning
Talk 1, 10:50 - 11:10
Kevin Mildau
specXplore: An interactive dashboard for flexible exploration of mass spectral data
Talk 2, 11:10 - 11:30
Markus Riedl
Comprehensive meta-analysis of the CHO coding transcriptome
Talk 3, 11:30 - 11:50
Roman Ufimov
ParalogWizard: A novel method for detecting and incorporating paralogues in phylogenetic tree inference from target enrichment data
Lunch break and Poster session
11:50 - 13:20
2nd session
13:20 - 15:00
Chairs: Christoph Bueschl, Markus Aigensberger
Keynote 13:20 - 14:00
Thomas Schwarzl
Bioinformatics binding site detection of enigmatic RNA-binding enzymes
Talk 4, 14:00 - 14:20
Markus Aigensberger
Comparison of untargeted metabolomics processing software, data treatment, & statistical analysis methods
Talk 5, 14:20 - 14:40
Jonathan Samson
TopN Simulator: A software tool for the optimization of data dependent analysis settings
Talk 6, 14:40 - 15:00
Katharina Munk
Holomics: an R Shiny application for the holistic integration and analysis of multiple omics data
Coffee break and Poster session
15:00 - 15:30
3rd session
15:30 - 17:10
Chairs: Guenter Brader, Katharina Munk
Keynote 15:30 - 16:10
Ulrik Sundekilde
Using multiomics to gain insight into the mother-milk-infant triad
Talk 7, 16:10 - 16:30
Muhammad Ahmad
Integrating high-throughput phenotyping with transcriptomics and metabolomics to gain insight into drought stress responses in Norway spruce seedlings
Talk 8, 16:30 - 16:50
Michael Santangeli
Deciphering the interplay of plant gene expression, root exudation and soil microbial community in the rhizosphere of field-grown maize across the vegetation period
Talk 9, 16:50 - 17:10
Anela Tosevska
Cytokine-derived transcriptomic signatures, cellular crosstalk and synovial pathotypes in rheumatoid arthritis
Poster prize and closing remarks
17:10 - 17:25
Closing reception with snacks and drinks
17:30 - open end
Book of Abstracts
Please find the book of abstracts here.
Organizing Committee
Franz Berthiller, Guenter Brader, Christoph Bueschl, Angelika Weiler, Birgit Herbinger, Alexandra Parich, Rainer Schuhmacher
Scientific Committee
Franz Berthiller, Guenter Brader, Christoph Bueschl, Birgit Herbinger, Krunic Milica, Thomas Rattei, Rainer Schuhmacher