Screenshots
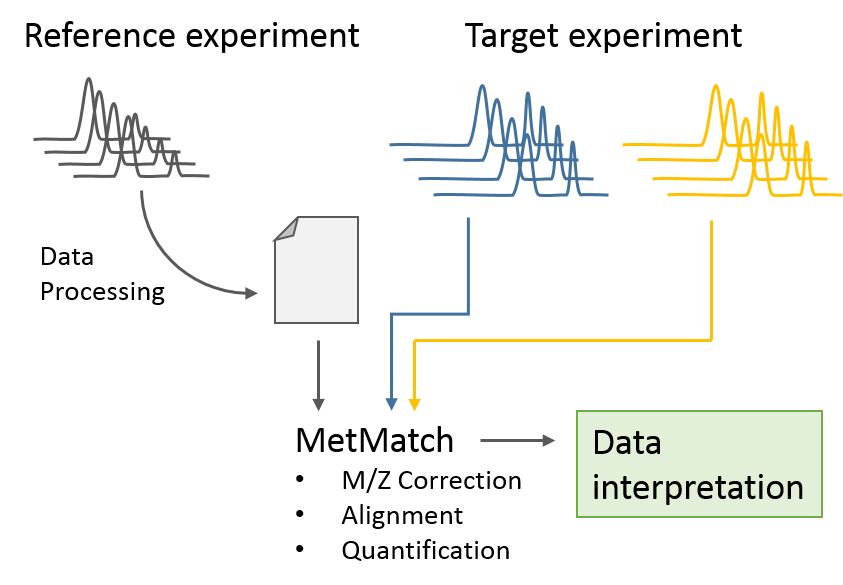
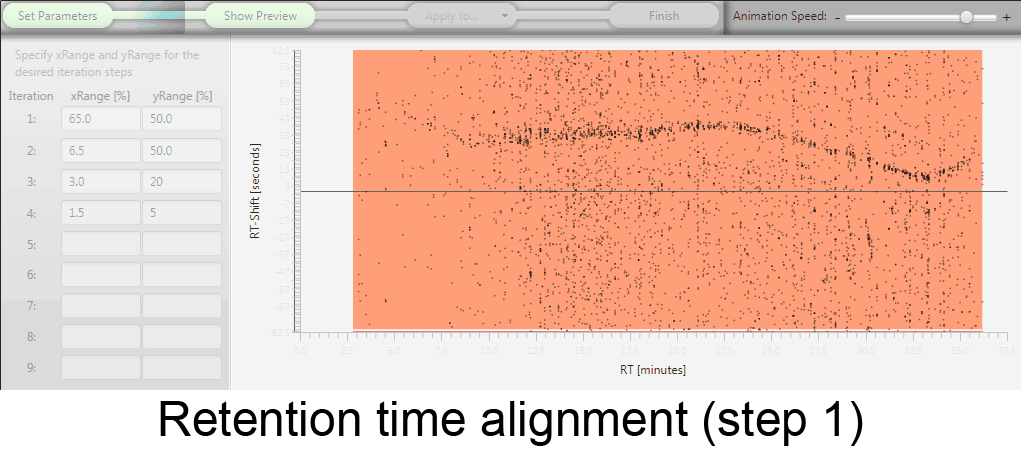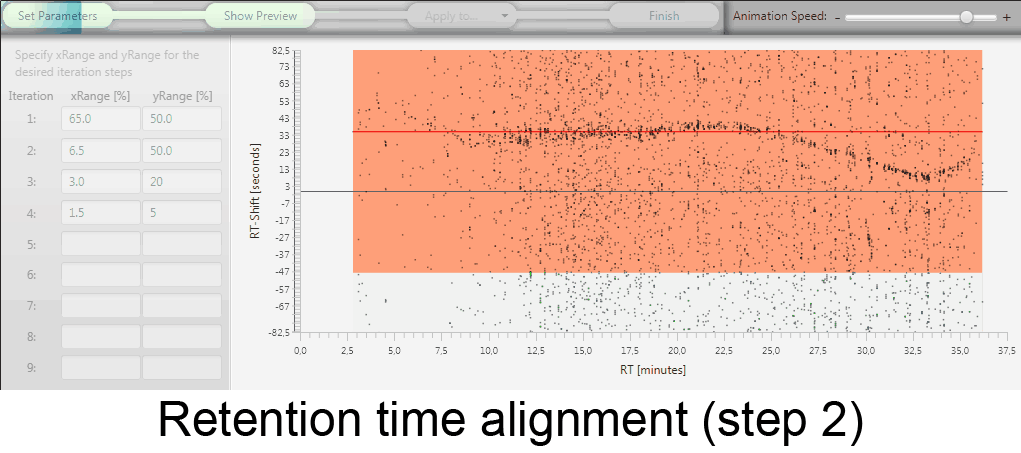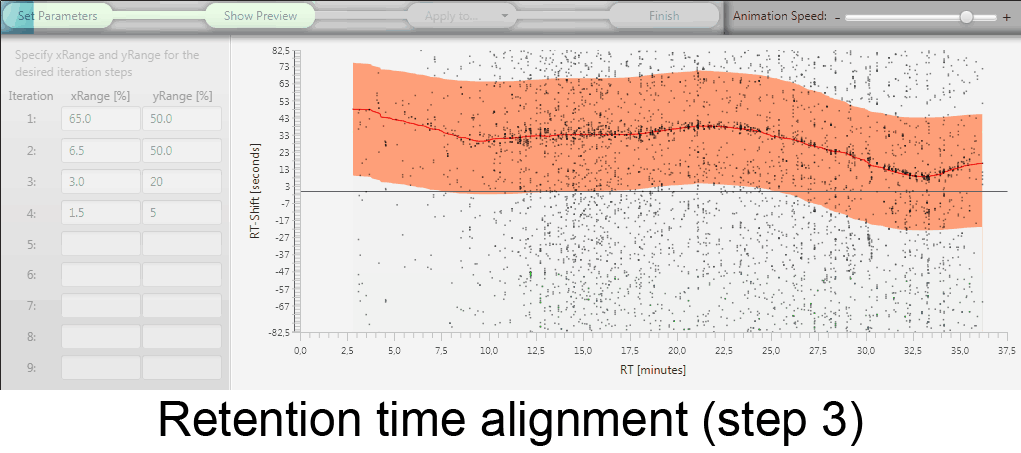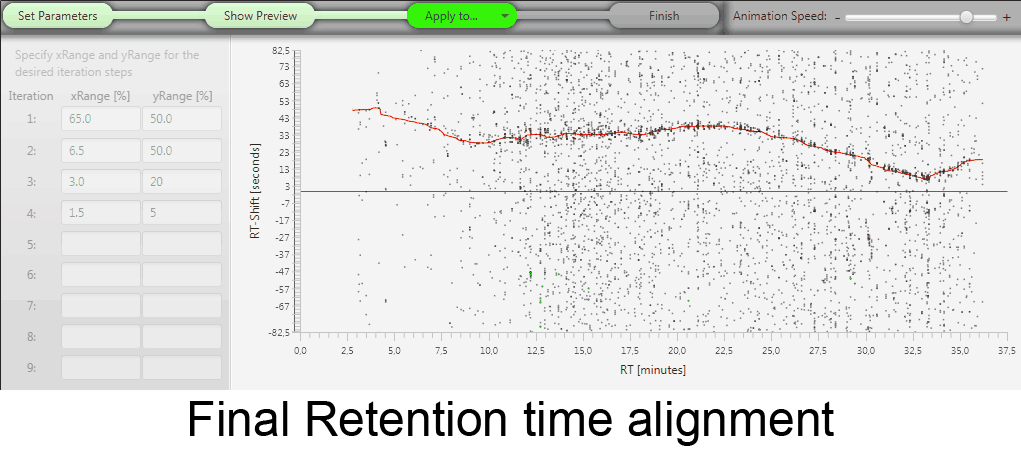
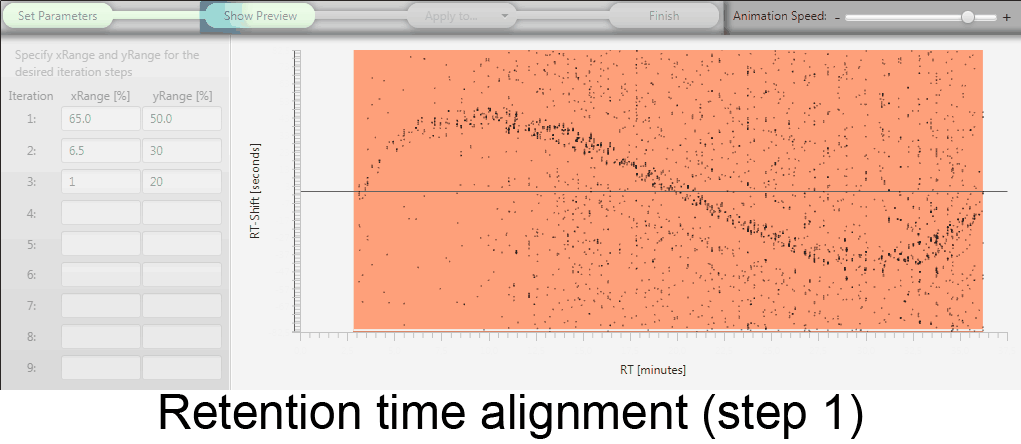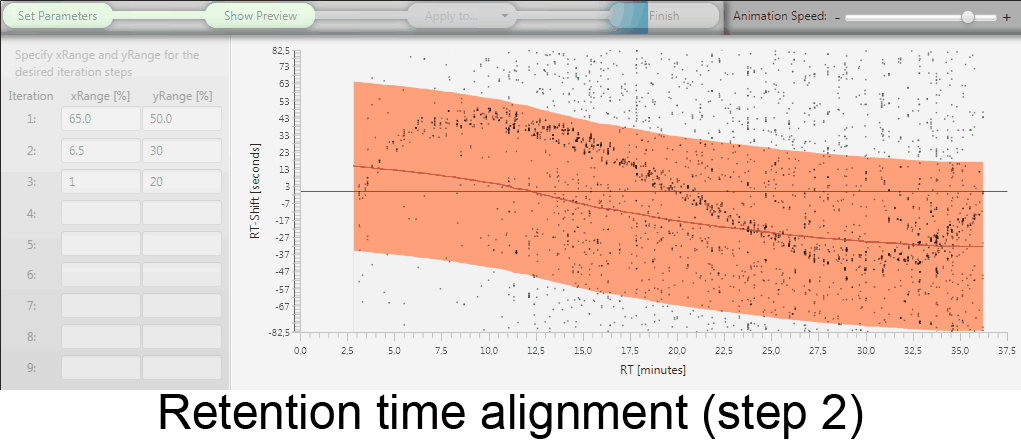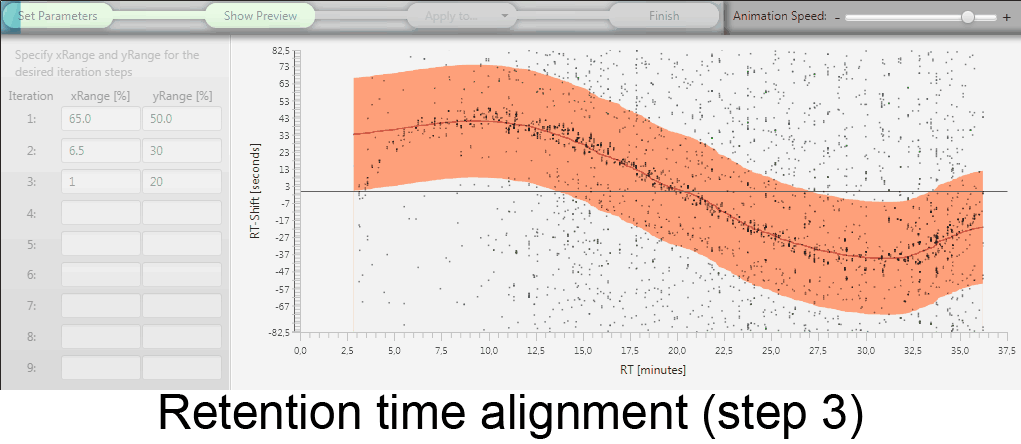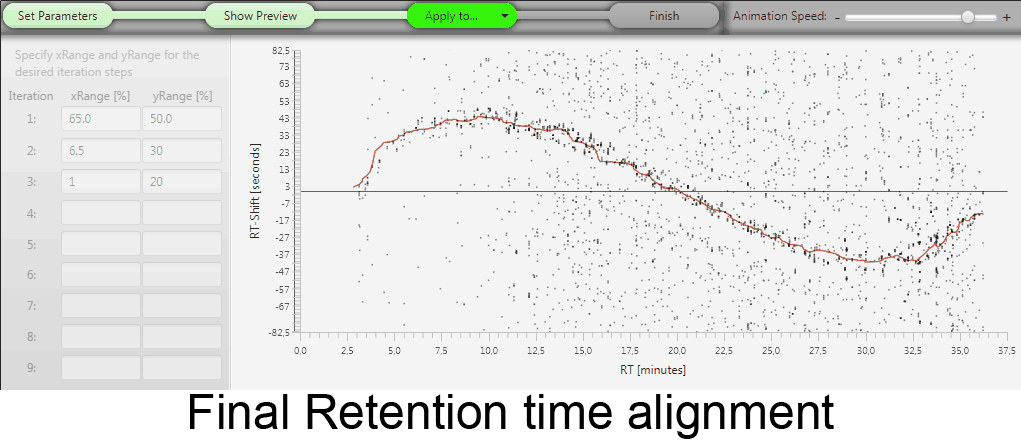
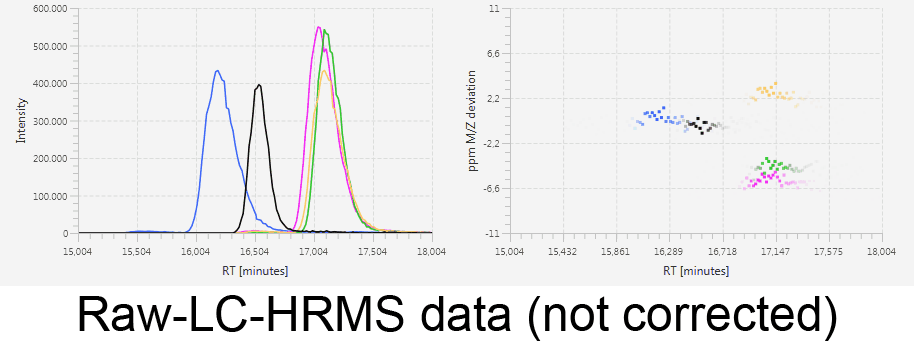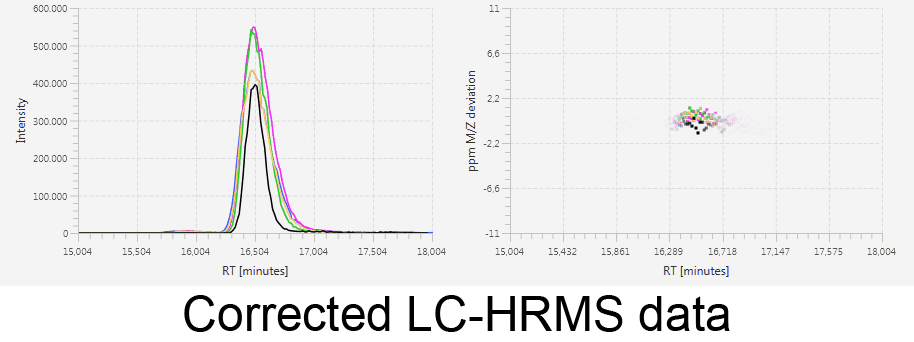
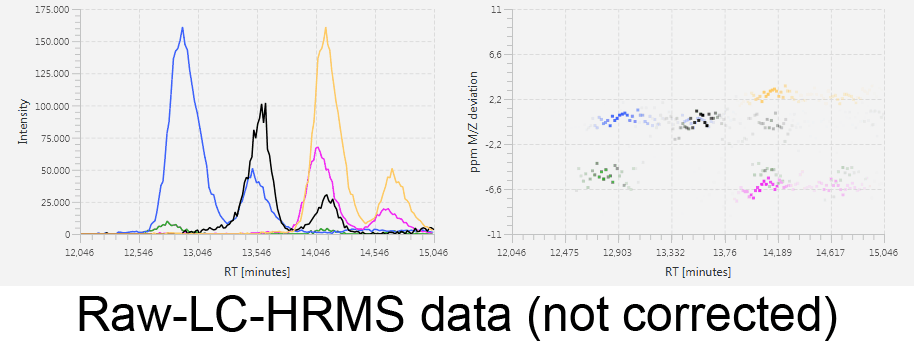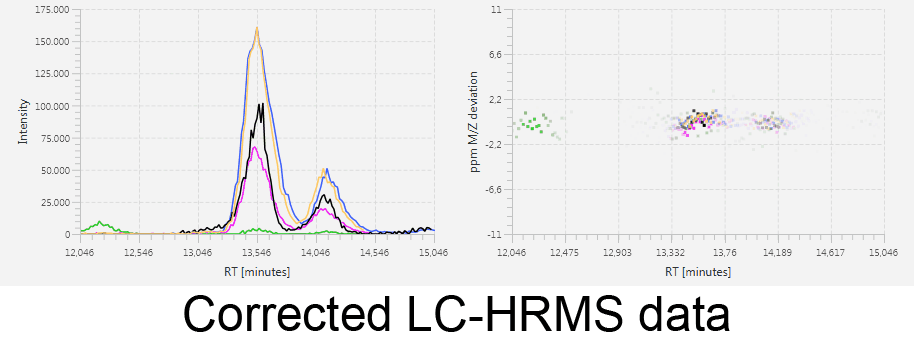
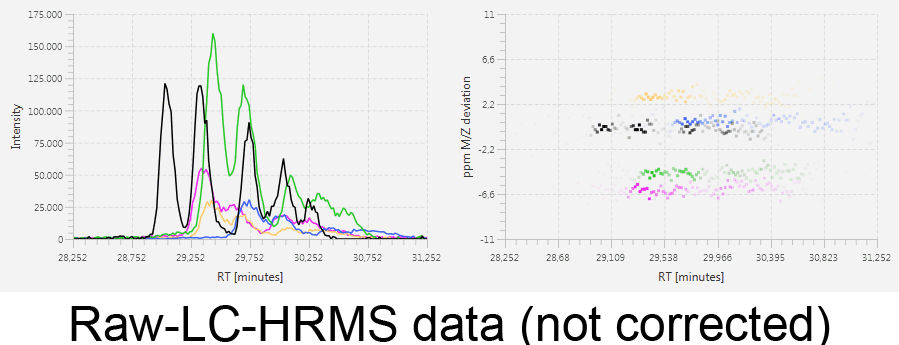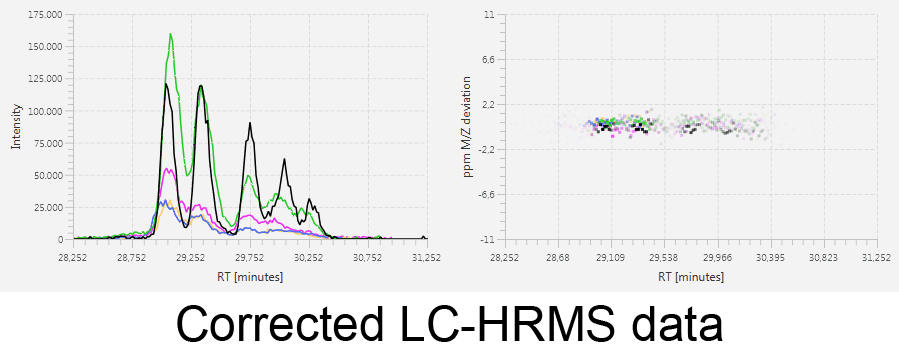
MetMatch is a software tool for the semi-automated comparison of different LC-HRMS chromatograms of untargeted metabolomics experiments. It provides methods to correct the raw-LC-HRMS data for m/z and retention time shifts relative to one or several preprocessed reference chromatograms.
MetMatch has been tested with modern LC-HRMS instruments including Thermo Fisher Orbitrap and Agilent QToF instruments. The software, associated documentation and sample datasets are avaialble free of charge and warranty.
Download the file and double-click it to start the installation process.
R will be automatically configurated upon installation of MetMatch. If not, please refer to the documentation for correctly configuring your system.
Documentation of MetExtract is included with the download or available here.






MetMatch is designed for the automated processing of LC-HRMS datasets. It calculates, aligns and corrects putative m/z and retention time deviation shifts between different measurement sequences or batches. Additionally, MetMatch is also capable of addressing the problem of different ion formation, which can be observed after a change of elution solvents.
MetMatch requires at least two experiments:
A reference dataset, which has to be processed with a software tool to detect the biologically relevant features, is used as a template.
One or more target datasets are then aligned to the reference dataset using the generated feature list.

MetMatch uses the open-data format mzXML to read and write LC-HRMS data. Thus, it virtually supports all different kinds of LC-HRMS instruments and vendors and has been tested with datasets obtained from an LTQ Orbitrap XL instrument (Thermo Fisher Scientific) and a 6550 QTOF instrument (Agilent technologies).
Sample data from the publication used for the simulated datasets.
The provided dataset consists of the LC-HRMS sample used for creating the simulated datasets in the publication of the software. The following different files are required for reproducing these results:
Chromatogram (36 MB)
This file is the LC-HRMS sample used for generating the simulated reference list and alignment of the data to this lists. It was recorded on an LTQ Orbitrap XL instrument in the positive ESI mode.
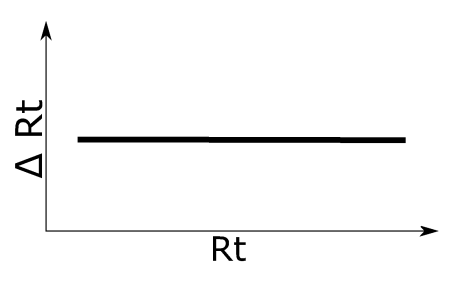
Simulation 1
A constant Rt shift was assumed. See left figure for an illustration of the applied retention time shift.
Simulation 2
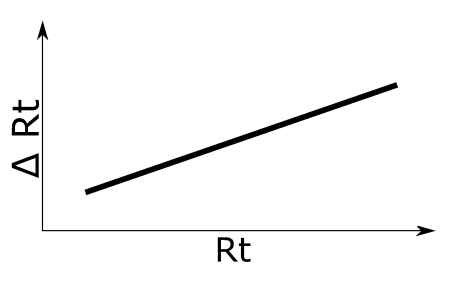
Monotonically increasing linear Rt shift. See left figure for an illustration of the applied retention time shift.
Simulation 3
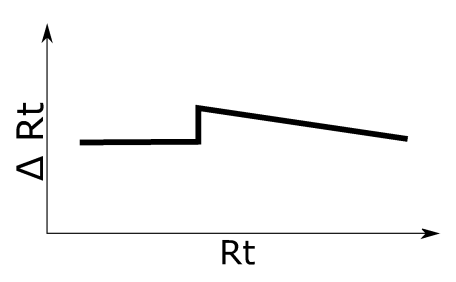
Discontinuous Rt shift, as may e.g. happen by a defective LC-pump. See left figure for an illustration of the applied retention time shift.
Simulation 4
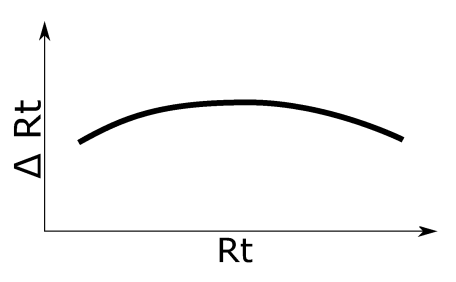
Non-linear Rt shift without inflection. See left figure for an illustration of the applied retention time shift.
Simulation 5
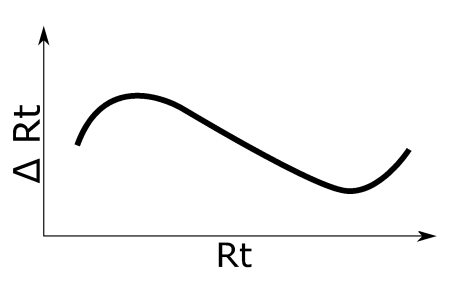
Non-linear Rt shift with inflection. See left figure for an illustration of the applied retention time shift.
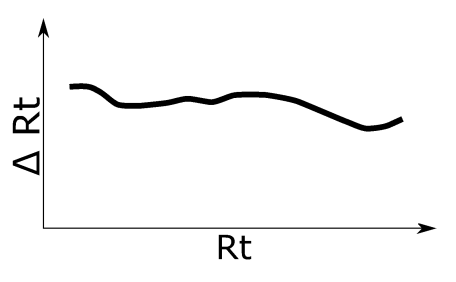
Simulation 6
Complex non-linear Rt shift based on the type of Rt shift observed in the real-world wheat experiment. See left figure for an illustration of the applied retention time shift.
Documentation of the MetMatch software is available as a part of the software or can be downloaded here
The source code of MetMatch is available on https://github.com/S-T-K/MetMatch
Sample applications of MetMatch can be found in:
Other software tools for untargeted metabolomics are: